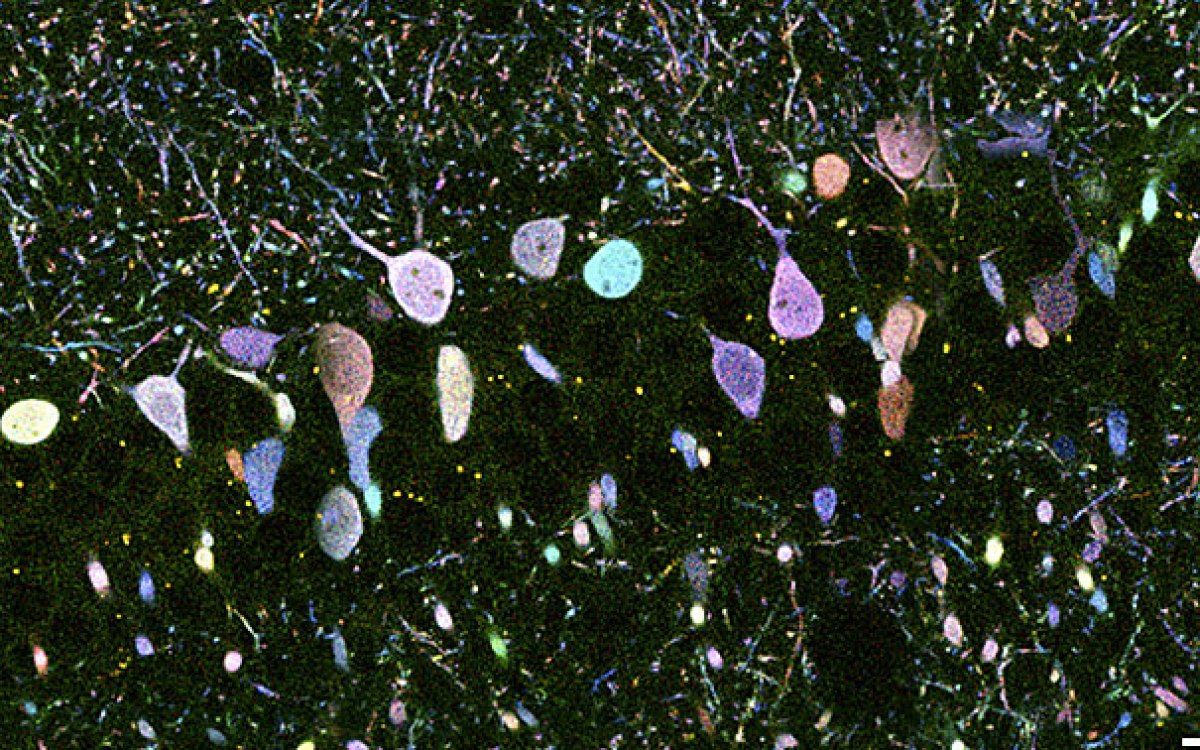
Harvard researchers examined 200 genes implicated in blinding diseases and found that some, particularly those associated with macular degeneration, are expressed selectively in foveal cells. The fovea is a small, specialized area in the retina that makes sharp central vision possible.
Harvard file photo
Focusing on the fovea
First cellular atlas could prove valuable resource in study of blinding diseases
For decades, scientists have turned to the retina of the humble laboratory mouse as an ideal model organism for understanding how neurons connect to form circuits in the brain. But as a model for vision and vision-related diseases, mice simply aren’t equipped.
The problem, said Joshua Sanes, the Jeff C. Tarr Professor of Molecular and Cellular Biology and director of the Center for Brain Science, is that they lack a fovea — a small, specialized area in the retina that makes sharp central vision possible. Among mammals, only primates have a fovea.
The fovea has been well-studied for decades. Researchers have demonstrated both functional and structural specialization in foveal cells, but the mechanisms that give rise to the differences between the fovea and the peripheral retina have remained a mystery.
To begin unraveling that mystery, a team of researchers led by Sanes applied high-throughput genetic sequencing methods to create the first cellular atlas of the primate retina. They found that while the fovea and peripheral retina share most of the same cell types, cells are found in different proportions in each region, and many show different gene-expression patterns.
The study, Sanes said, offers an important foundation for researchers to build on as they seek to understand how vision works in primates, including humans, and how it can be disrupted by disease. The work is described in a paper published in Cell on Feb. 21.
To create the cellular atlas, Sanes and his colleagues began with 165,000 cells collected from the retinas of macaques, about half of them foveal and half peripheral. The team used genetic tools to separate them into their various types, identifying 65 to 70 separate types in both parts of the eye, along with the genes each type expresses.
The results produced both good and bad news.
The good news, Sanes said, was that the researchers came up with ideas about what makes the fovea special. “About 90 percent of the cell types are shared,” Sanes said, “but what is more telling is that … [the two types] express a lot of different genes. We believe that looking at those genes will help us explain a lot of the functional difference between a cell in the fovea and one in the periphery.”

The bad news was that when the team tried to find mouse equivalents of the main cell type that sends a message from the fovea to the brain — called the “midget ganglion cell” for its small size — they largely came up empty.
“Unfortunately, ‘midgets’ are the vast majority of foveal ganglion cells,” Sanes said. “We’d hoped we would find the mouse equivalent of those cells … so we could study them using all the tools we’ve already developed, but we didn’t. So that was a disappointment.”
Armed with their cellular atlas, Sanes and his colleagues turned to nearly 200 genes implicated in blinding diseases and found that some, particularly those associated with macular degeneration and diabetic macular edema, are expressed selectively in foveal cells — a tantalizing clue for why such diseases primarily affect the fovea: The macula is a slightly larger region of the retina with the fovea at its center, so “this pattern makes sense,” said Sanes.
“For example, we found one macular degeneration susceptibility gene is expressed at substantially higher levels in foveal rods and cones than in peripheral rods and cones,” Sanes said. “That may be related to the fact that it’s a macular disease. Similarly for diabetic macular edema — we found two susceptibility genes expressed at higher levels in the blood vessels in the fovea than in the periphery.” Macular degeneration and diabetic macular edema are among the leading causes of blindness in the U.S. and the world.
“We also took those genes back to the mouse cellular atlas,” Sanes continued, “and in many cases they either weren’t expressed or were expressed in different cells. That suggests that, for at least some of these diseases, looking at how a gene is acting in mice may not be very telling, because it may not be expressed in the same cells.”
Going forward, Sanes said, the cellular atlas could serve as a valuable resource both for studies focused on specific diseases and for basic science questions.
“On the disease side, just by having this knowledge we may be able to think about better treatments,” Sanes said. “This could also be a foundation for looking at pathological material. We are collaborating with people who have banked eyes over the years for various purposes, which means we could begin to test hypotheses — so if we think perhaps people with glaucoma are missing a certain cell type or the expression of a gene is too low or too high, we can test that.
“On the basic research side,” he continued, “we can take a gene that we may have reason to think could play a role in the physiological differences in the cells and put it in a mouse cell and see if it changes the properties of that cell. This will give us a big head start as we look to answer these kinds of questions.”
This research was supported with funding from the National Eye Institute (part of the National Institutes of Health), the Broad Institute, and the BrightFocus Foundation.