New study finds that DNA configuration help generate diverse antibodies
New studies have provided a major advance in knowledge of how diverse antibodies are generated. Illustration courtesy of Boston Children’s Hospital
We need a variety of antibody types to help fight off invading foreign pathogens and our genome is exquisitely tuned to produce them to meet emerging needs. A new study finds that not just our DNA, but its configuration and packaging, help us generate diverse antibodies.
Our DNA strands are organized, together with certain proteins, into a packaging called chromatin.
In a paper published in the journal Nature, researchers from the laboratory of Frederick Alt of the Program in Cellular and Molecular Medicine (PCMM) at Boston Children’s Hospital reveal insights into a new mechanism of chromatin regulation — changing the configuration of our DNA and its packaging — and how that influences antibody formation and gene regulation in general.
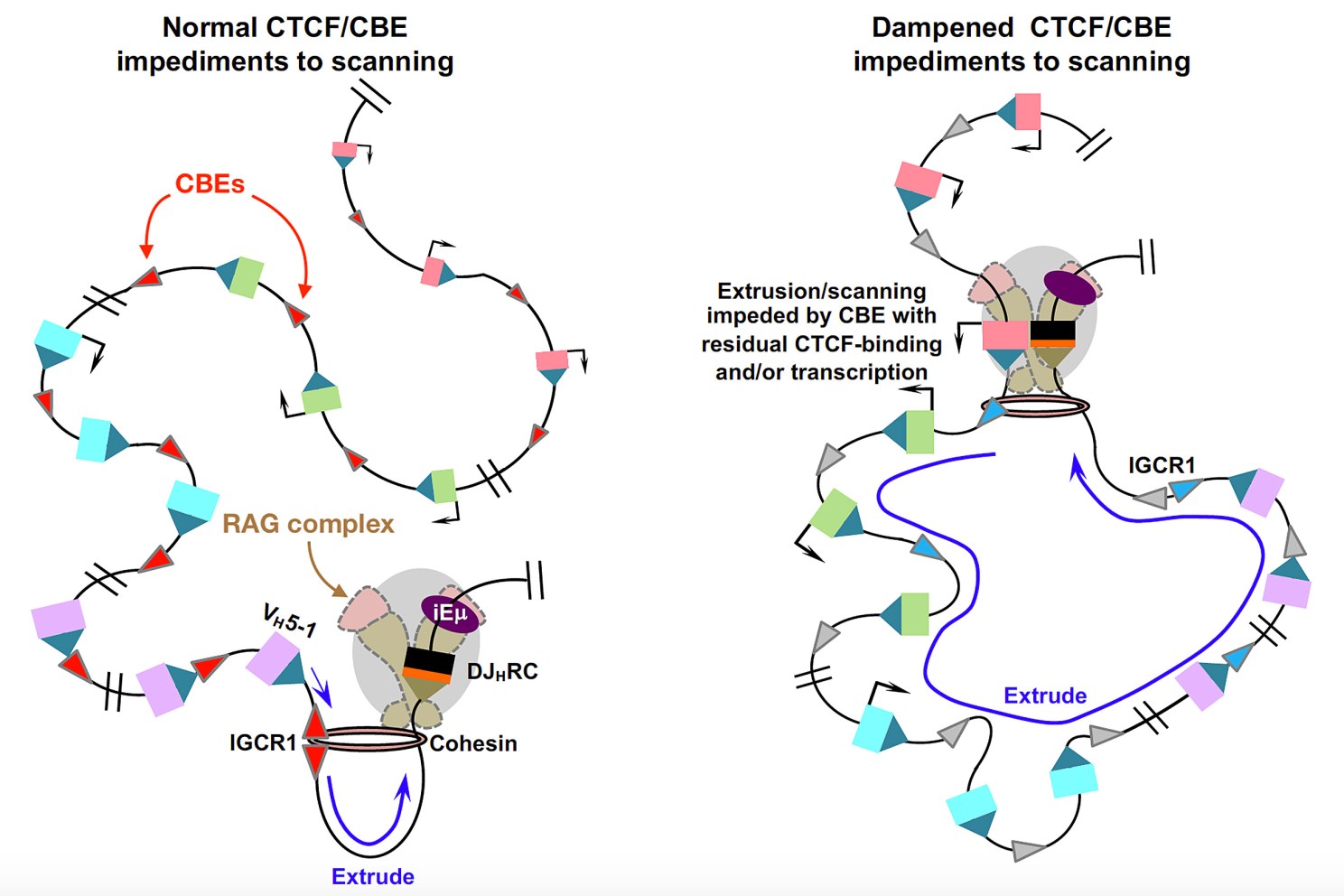
The team showed that cohesin, a protein complex with a major role in organizing chromatin, is a key player in forming new loops in chromatin across long antibody genes. As the Alt lab showed earlier, this loop extrusion is essential for the process of V(D)J recombination — which bring bits of genetic code together from long distances to form new antibodies.
Cohesin is the ring leader
“There has been a lot of discussion about whether or not cohesin really is the protein complex that is responsible for this major process that organizes the mammalian genome into loops — loops that could be very important for all kinds of gene regulation, including antibody genes,” said Alt. These loop configurations physically position portions of genes that ought to work together in close proximity to one another, increasing their ability to work in tandem.
The process of cohesin-enabled loop extrusion stops only when cohesin encounters blockades called CTCF-binding elements (CBEs). CBEs help segregate the entire genome into well-defined loops.
Cohensin plus V(D)J recombination leads to diverse antibody types
For V(D)J recombination to occur, cohesin sits at one point on the DNA but somehow pulls past it hundreds of gene segments scattered across the chromatin, often from huge distances. That process leads to the generation of a large repertoire of antibodies.
“If we didn’t have diverse antibody repertoires, we would all be immunodeficient and might die, particularly in the face of the current pandemic,” said Alt.
Until now, it was not understood how our immune system cells find these different gene segments over long distances of the chromatin, pick them out, and put them together to make very diverse antibody repertoires.
Rolling past roadblocks
The paper addresses another important mystery in chromatin biology: How does cohesin get past the CBE obstacles on the chromatin that naturally prevent loop formation over long distances?
“We found that shutting down the CBEs allows cohesin to move large regions of chromatin to bring gene segments together,” said Zhaoqing Ba of the Alt lab, first author on the paper. “This ultimately allows the V(D)J recombination complex to gather and assemble these important gene segments that are required to have very broad antibody repertoires.”
A new form of gene regulation
Their research has larger implications beyond production of antibody diversity. It points to potential new mechanisms explaining long-range gene regulation in both health and disease.
“Silencing CBE-blocking activity and allowing cohesin to pull long stretches of chromatin together may be important for regulation of many types of genes,” said Alt. “Our study provides an important step for understanding a new form of gene regulation at the level of chromatin architecture and chromatin biology, which could be very important for all forms of gene regulation.”




