
An accompanying smartphone app analyzes the fluorescent readout on miSHERLOCK, giving users a clear “Positive” or “Negative” result.
Images courtesy of Wyss Institute at Harvard University
New test detects COVID-19 variants in your spit
Low-cost device lets consumers check themselves for multiple strains of SARS-CoV-2 at home
With the delta variant wreaking havoc on unvaccinated populations, the ability to quickly diagnose and track emerging variants of the virus is crucial. Researchers have now created a simple, inexpensive, CRISPR-based diagnostic test that allows users to test themselves for multiple variants of the SARS-CoV-2 virus at home, using just a sample of their saliva.
Developed by researchers at the Wyss Institute for Biologically Inspired Engineering at Harvard University, the Massachusetts Institute of Technology, and several Boston-area hospitals, the diagnostic device, called Minimally Instrumented SHERLOCK (miSHERLOCK), is easy to use and provides results that can be read and verified by an accompanying smartphone app within one hour.
miSHERLOCK successfully distinguished between three different variants of SARS-CoV-2 in experiments, and can be rapidly reconfigured to detect additional variants like delta. The device can be assembled using a 3D printer and commonly available components for about $15, and re-using the hardware brings the cost of individual assays down to $6 each.
“miSHERLOCK eliminates the need to transport patient samples to a centralized testing location and greatly simplifies the sample preparation steps, giving patients and doctors a faster, more accurate picture of individual and community health, which is critical during an evolving pandemic,” said co-first author Helena de Puig, a postdoctoral fellow at the Wyss Institute and MIT.
The diagnostic device is described in a paper published Friday in Science Advances.
From supply chain to SHERLOCK
As an instructor in pediatrics at Boston Children’s Hospital with a specialization in infectious diseases, co-first author Rose Lee has been working on the front lines of the COVID-19 pandemic for over a year. Her experiences in the clinic provided inspiration for the project that would ultimately become miSHERLOCK.
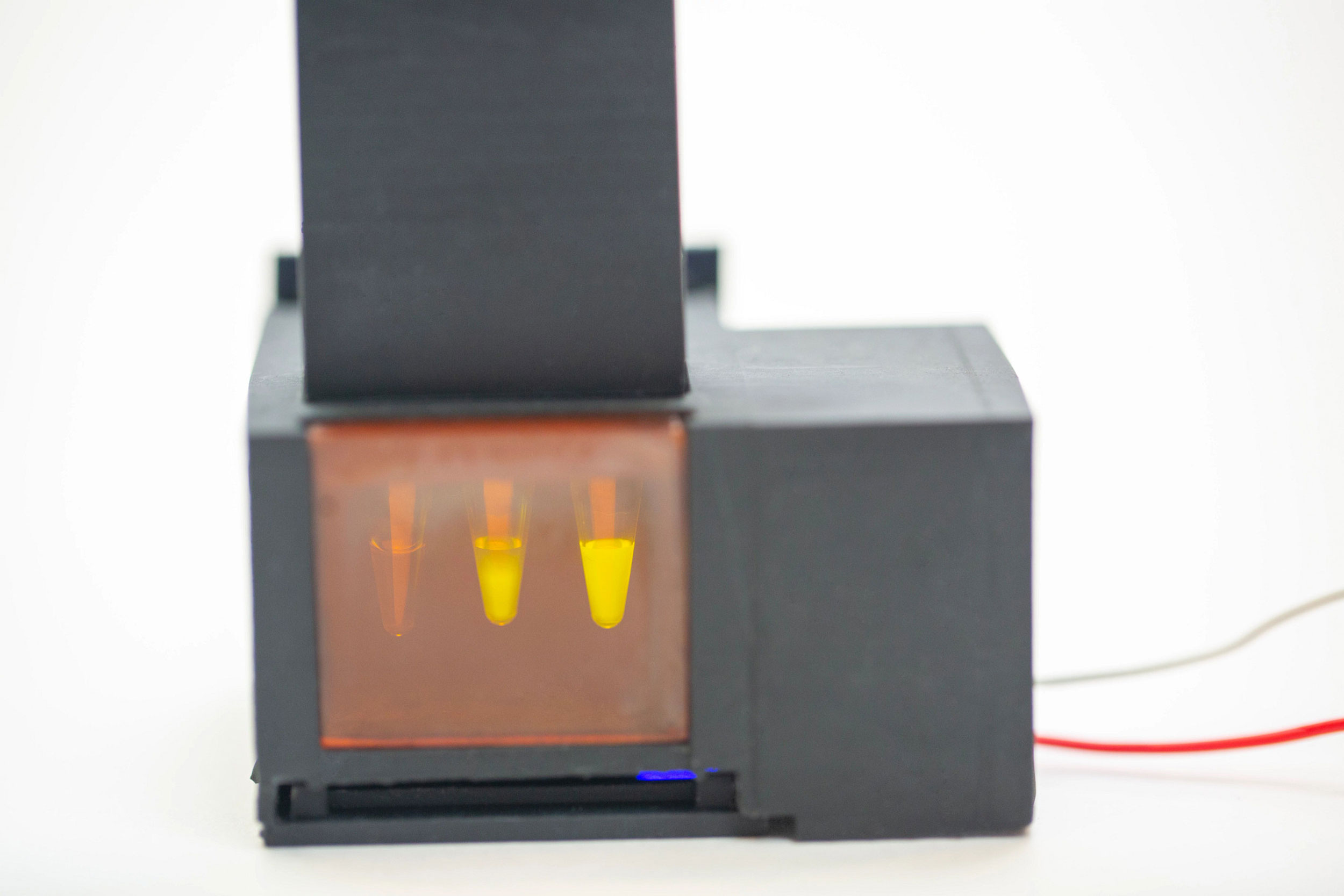

The device produces a fluorescent signal in response to the presence of SARS-CoV-2 viral RNA in a patient’s saliva. The four first authors of the miSHERLOCK paper, pictured right to left: Devora Najjar, Rose A. Lee, Helena de Puig, and Xiao Tan.
“Simple things that used to be ubiquitous in the hospital, like nasopharyngeal swabs, were suddenly hard to get, so routine sample processing procedures were disrupted, which is a big problem in a pandemic setting,” said Lee, who is also a visiting fellow at the Wyss Institute. “Our team’s motivation for this project was to eliminate these bottlenecks and provide accurate diagnostics for COVID-19 with less reliance on global supply chains, and also accurately detect the variants that were starting to emerge.”
For the SARS-CoV-2 detection piece of their diagnostic, the group turned to a CRISPR-based technology created in the lab of Wyss Core Faculty member and senior paper author Jim Collins, called “specific high sensitivity enzymatic reporter unlocking” (SHERLOCK). SHERLOCK makes use of CRISPR’s “molecular scissors” to snip DNA or RNA at specific locations, with an added bonus: this specific type of scissors also cuts other pieces of DNA in the surrounding area, allowing it to be engineered with nucleic acid probe molecules to produce a signal indicating that the target has been successfully cut.
The researchers created a SHERLOCK reaction designed to cut SARS-CoV-2 RNA at a specific region of a gene called Nucleoprotein that is conserved across multiple variants of the virus. When the molecular scissors — an enzyme called Cas12a — successfully bind to and cut the nucleoprotein gene, single-stranded DNA probes are also cut, producing a fluorescent signal. They also created additional SHERLOCK assays designed to target a panel of viral mutations in Spike protein sequences that represent three SARS-CoV-2 genetic variants: Alpha, Beta, and Gamma.
Armed with assays that could reliably detect viral RNA within the accepted concentration range for FDA-authorized diagnostic tests, the team next focused their efforts on solving what is arguably the most difficult challenge in diagnostics: sample preparation.
The team chose to use saliva rather than nasopharyngeal swab samples as their collection method, because it’s easier for users to collect saliva and studies have shown that SARS-CoV-2 is detectable in saliva for a greater number of days post-infection. But unprocessed saliva presents challenges of its own: It contains enzymes that degrade various molecules, producing a high rate of false positives.
The researchers developed a novel technique to solve that problem. First, they added two chemicals called DTT and EGTA to saliva and heated the sample to 95°C for 3 minutes, which eliminated the false-positive signal from the untreated saliva and sliced open any viral particles. They then incorporated a porous membrane that was engineered to trap RNA on its surface, which could finally be added directly to the SHERLOCK reaction to generate a result.
Spit, wait, scan
To integrate the saliva sample prep and the SHERLOCK reaction into one diagnostic, the team designed a simple battery-powered device with two chambers: a heated sample prep chamber, and an unheated reaction chamber. A user spits into the sample prep chamber, turns on the heat, and waits three to six minutes for the saliva to be wicked into the filter. The user removes the filter and transfers it to the reaction chamber column, then pushes a plunger that deposits the filter into the chamber and punctures a water reservoir to activate the SHERLOCK reaction. Fifty-five minutes later, the user looks through the tinted transilluminator window into the reaction chamber and confirms the presence of a fluorescent signal. They can also use an accompanying smartphone app that analyzes the pixels being registered by the smartphone’s camera to provide a clear positive or negative diagnosis.
The researchers tested their diagnostic device using clinical saliva samples from 27 COVID-19 patients and 21 healthy patients, and found that miSHERLOCK correctly identified COVID-19-positive patients 96 percent of the time and patients without the disease 95 percent of the time. They also tested its performance against the Alpha, Beta, and Gamma SARS-CoV-2 variants by spiking healthy human saliva with full-length synthetic viral RNA containing mutations representing each variant, and found that the device was effective across a range of viral RNA concentrations.
“One of the great things about miSHERLOCK is that it’s entirely modular. The device itself is separate from the assays, so you can plug in different assays for the specific sequence of RNA or DNA you’re trying to detect,” said co-first author Devora Najjar, a research assistant at the MIT Media Lab and in the Collins Lab. “Assays for new targets can be created in about two weeks, enabling the rapid development of tests for new variants of COVID-19 and other diseases.”
Ready for the real world
The miSHERLOCK team created their device with low-resource settings in mind, as the pandemic has brought to light the vast inequalities in health care access that exist between different parts of the world.
“When the miSHERLOCK project started, there was almost no SARS-CoV-2 variant monitoring happening. We knew that variant tracking was going to be incredibly important when evaluating the long-term effects of COVID-19 on local and global communities, so we pushed ourselves to create a truly decentralized, flexible, user-friendly diagnostic platform,” said Collins, who is also the Termeer Professor of Medical Engineering & Science at MIT. “By solving the sample prep problem, we’ve ensured that this device is virtually ready for consumers to use as-is, and we’re excited to work with industrial partners to make it commercially available.”
This research was supported by the Wyss Institute for Biologically Inspired Engineering at Harvard University, the Paul G. Allen Frontiers Group, the Harvard University Center for AIDS Research (an NIH-funded program that is supported by the following NIH co-funding and participating institutes and centers: NIAID, NCI, NICHD, NIDCR, NHLBI, NIDA, NIMH, NIA, NIDDK, NINR, NIMHD, FIC, OAR), The Burroughs-Wellcome American Society of Tropical Medicine and Hygiene, an American Gastroenterological Association Takeda Pharmaceutical Research Scholar Award, and an MIT-TATA Center fellowship.
This story has been edited for length. For the full release, visit the Wyss Institute website.




