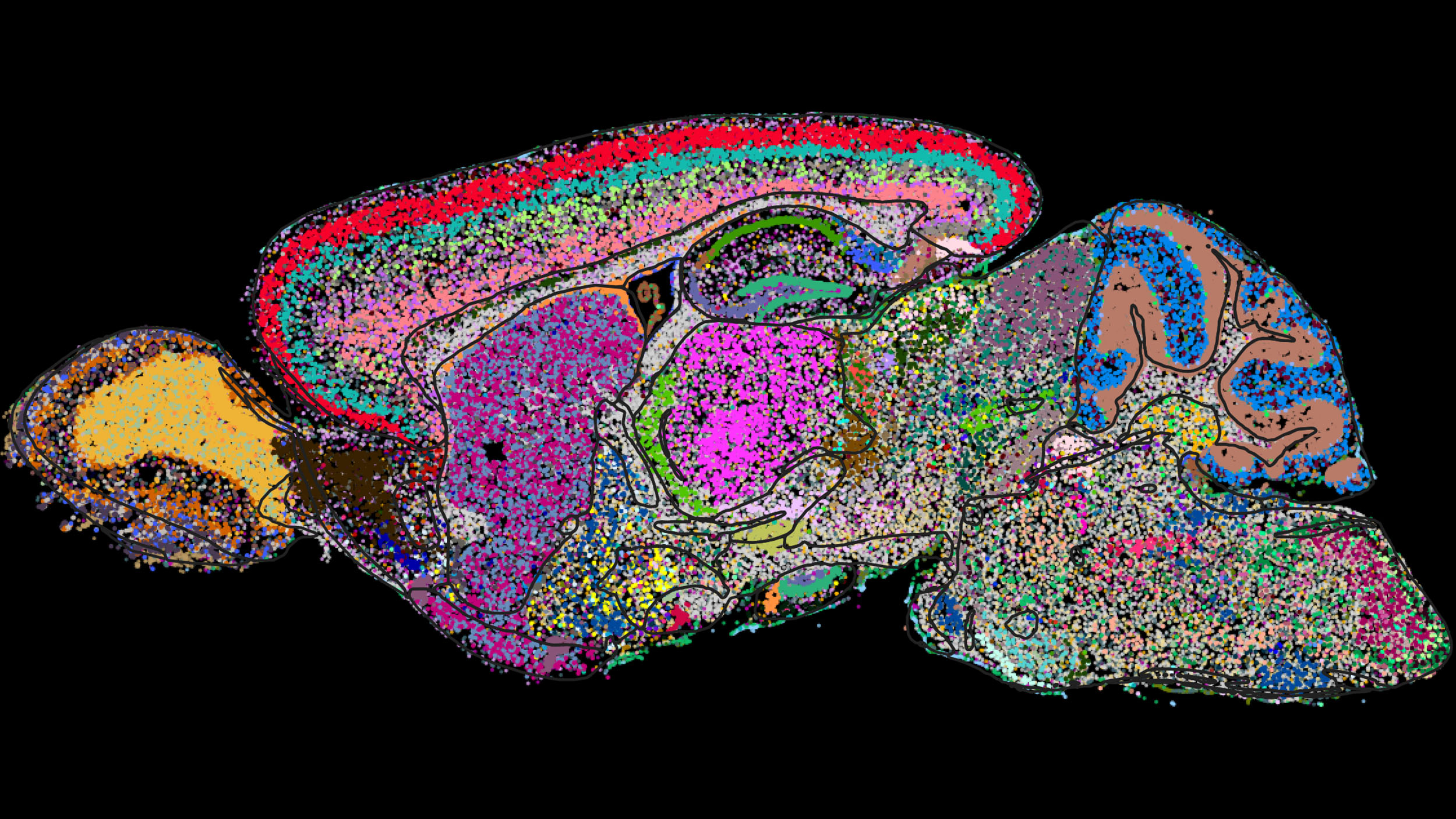
Cell atlas of the mouse brain determined by a genome-scale imaging technology called MERFISH. Shown is one sagittal section with cells colored by category.
Image courtesy of Xiaowei Zhuang
Demystifying a mammal’s brain, cell by cell
Harvard-led team helps create first molecular map for national neuroscience study
A group of scientists, including several at Harvard, have dived deeper into the mammalian brain than ever before by categorizing and mapping at the molecular level all of its thousands of different cell types.
The researchers reported their work in Nature, through a series of 10 papers — six with Harvard affiliations. It’s part of the National Institutes of Health’s Brain Research Through Advancing Innovative Neurotechnologies initiative, which so far has focused on mice; future phases will shift to humans and other primates.
Mammal brains house billions of cells, each defined by the genes they express. This complexity is why true understanding of many brain functions, including molecular mechanisms that underlie neurological diseases, remains so elusive.
To create the first molecularly defined cell atlas of the whole mouse brain, a team led by Harvard’s Xiaowei Zhuang identified and spatially mapped thousands of unique cell types, most of which had never previously been characterized.
“We identified 5,000 transcriptionally distinct cell populations,” said Zhuang, the David B. Arnold Professor of Science and a Howard Hughes medical investigator. “Suffice it to say that the level of diversity we identified is really extraordinary.”
The brain-wide atlas of cell types cataloging cells, their distribution, and interactions could serve as a starting point for scientists studying certain brain functions or diseases. Someday the basic outlines of the atlas could be applied to the human brain, 1,000 times larger than the mouse brain.
“It gives me real excitement to see things that were not visible before. I am also thrilled when our technology is used by so many labs,” said Zhuang, referring to Multiplexed Error-Robust Fluorescence in situ Hybridization (MERFISH), a genome-scale imaging technology developed in her lab.
Xiaowei Zhuang (center) in the lab with team Aaron Halpern (from left), Won Jung, Meng Zhang, and Xingjie Pan.
Niles Singer/Harvard Staff Photographer
In collaboration with scientists at the Allen Institute for Brain Science, Zhuang and her team used MERFISH together with single-cell RNA sequencing data to not only identify each cell type, but to image them in situ. Their work provides new information about the molecular signatures of these cell types, as well as where they are located in the brain. The result is a stunningly detailed picture of the mouse brain’s full complement of cells, their gene-expression activity, and their spatial relationships.
In their Nature paper, the researchers used MERFISH to determine gene-expression profiles of approximately 10 million cells by imaging a panel of 1,100 genes, selected using single-cell RNA sequencing data provided by Allen Institute collaborators.
Retina findings could boost glaucoma research
In a separate paper in the Nature series, Joshua Sanes, the Jeff C. Tarr Professor of Molecular and Cellular Biology, co-led a team that captured new insights into the evolutionary history of the vertebrate retina.

Joshua Sanes.
Photo by Rick Friedman
A part of the brain encased in the eye, the retina boasts complex neural circuits that receive visual information, which they then transmit to the rest of the brain for further processing. The retina is functionally very different from species to species — for example, human hunter-gatherers evolved sharp daytime vision, whereas mice possess better night vision than humans do; some animals see in color, while others see predominantly in black and white.
But at molecular levels, how different are retinas, really? Sanes, in collaboration with researchers at the University of California, Berkeley, and the Broad Institute, performed a new comparative analysis of retinal cell types across 17 species, including humans, fish, mice, and opossums. Using single-cell RNA sequencing, which allowed them to differentiate types of retinal cells by their genetic expression profiles, the researchers’ findings upended some long-held views about how certain species’ visual systems evolved.
One striking discovery involved so-called “midget retinal ganglion cells,” which, in humans, carry 90 percent of the information from the eye to the brain. These cells give humans their fine-detail vision, and changes to them are associated with eye diseases such as glaucoma. No related cells had ever been found in mice, so they had been assumed to be unique to primates.
In their analysis, Sanes and team identified for the first time clear relatives of midget retinal ganglion cells in many other species, including mice, albeit in much smaller proportions. Since mice are a common model animal to study glaucoma, being able to pinpoint these cells is a potentially crucial insight.
“I think we can make a very compelling case that if you want to study these important human retinal ganglion cells in a mouse, these are the cells you want to be studying,” Sanes said.
Other Harvard-affiliated researchers, at Harvard Medical School, Boston Children’s Hospital, and the Broad, also contributed findings to the NIH’s cell census network, including a molecular cytoarchitecture of the adult mouse brain, and a transcriptomic taxonomy of mouse brain-wide spinal projecting neurons.





