
Wyss postdoctoral fellow Amir Bein checks a batch of human organ chips in the lab.
Credit: Wyss Institute at Harvard University
Wyss Institute to accelerate drug testing for COVID treatment
Multicenter collaboration aims to rapidly identify FDA-approved drugs that can prevent or treat COVID-19 infections
The Wyss Institute and the U.S. Defense Advanced Research Projects Agency (DARPA) have signed an agreement worth up to $16 million over the next year to use Wyss technologies to identify and test FDA-approved drugs that could be repurposed to prevent or treat COVID-19.
Specifically, DARPA will use the computational drug-discovery pipelines and human organ chip technologies developed by the Wyss Institute for Biologically Inspired Engineering at Harvard University. The team, led by Wyss Founding Director Donald Ingber, has already found multiple approved compounds that show promise against the SARS-CoV-2 virus that causes COVID-19.
Many more drugs and lead compounds are being tested in high-throughput cell-based assays with native CoV-2 virus in the lab of Matthew Frieman, associate professor of microbiology and immunology at the University of Maryland School of Medicine. The most promising drugs are being transferred to the lab of Benjamin tenOever at the Icahn School of Medicine at Mount Sinai for testing in COVID-19 animal models.
Human organ chip technology is also being installed in the Frieman and tenOever labs with equipment from the Wyss spinout Emulate, Inc., that will let them analyze the human response to COVID-19 infection in vitro.
“Over the past few years, the Wyss Institute has been building up its computational approaches to identify compounds as potential therapeutics and validate them using our human organ chip microfluidic culture technologies to validate them, but the emergence of COVID-19 has really galvanized us to quickly integrate all of our capabilities and bring full force to bear on that challenge,” said Ingber, who is also the Judah Folkman Professor of Vascular Biology at Harvard Medical School and Boston Children’s Hospital, and professor of bioengineering at the Harvard John A. Paulson School of Engineering and Applied Sciences.
“Our initial successes allowed us to obtain this new support from DARPA, which we hope will greatly accelerate the development of drugs that might be used to prevent the spread of disease in large populations, as this is precisely what is needed before we can all go back to something close to life as usual.”
In addition to identifying and testing compounds for potential use against the virus, the Wyss team has established relationships with the Beth Israel Deaconess Medical Center (BIDMC) and SUNY Downstate Medical Center, where they are collecting clinical specimens from COVID-19 patients and carrying out RNA analysis using the sequencing core at the Broad Institute of Harvard and MIT, which will then provide data that can be fed back into the institute’s computational discovery pipeline.
Computing a COVID-19 cure
There are no treatments or vaccines for the novel coronavirus because it is just that: novel. Treatment now largely consists of supportive care to give patients’ immune systems the best shot at overcoming the virus on their own, but many patients cannot survive the viral onslaught. To date, testing FDA-approved drugs to determine if they can be repurposed to treat COVID-19 has not been pursued in a careful and systematic way. As a result, there has been much speculation in the media regarding unproven and/or off-label use of approved medications as potential therapies.
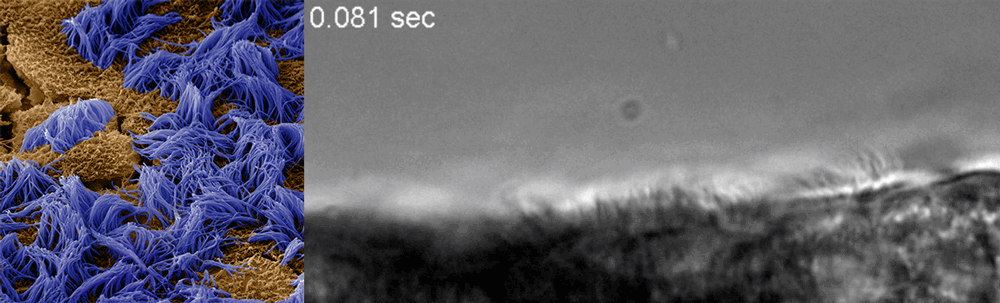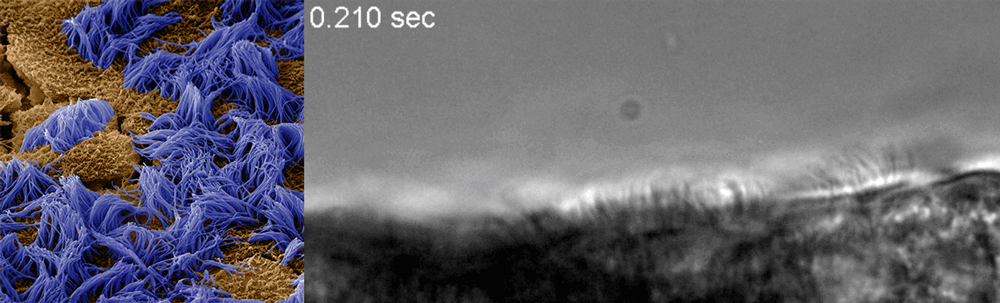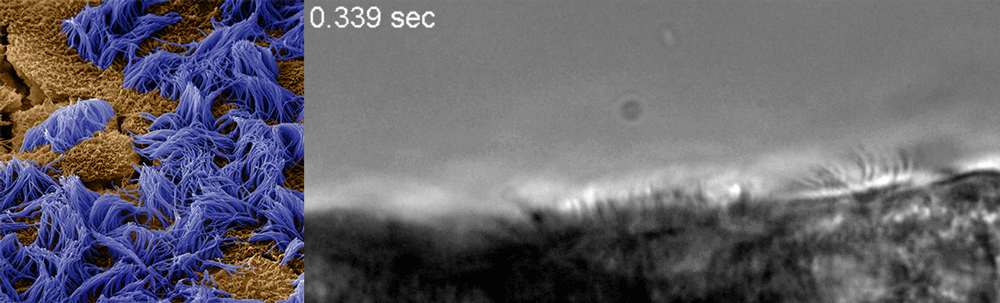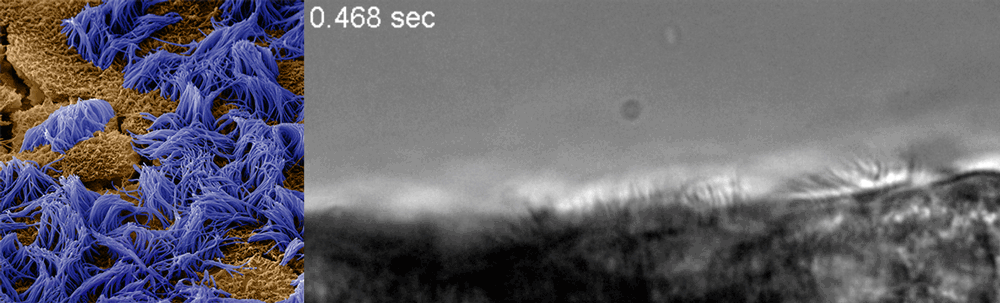
Ingber said he recognized that drug discovery efforts already underway at the Wyss could be adapted to this end, and created a Coronavirus Therapeutics Project Team. Composed of members with diverse skill sets, from analytical chemistry to machine learning to pathophysiology and virology, the team’s work now focuses nearly exclusively on finding and testing drugs that could potentially treat COVID-19.
“The processes that we’re now putting in place are bringing the Wyss up to par with the world’s leading drug-development companies and institutions, and we have the potential to even go beyond the current standard because we’re using novel approaches,” said Ken Carlson, a senior staff scientist at the Wyss and lead of the Coronavirus Therapeutics Project Team.
The pandemic has accelerated the pace of that effort by many months, added Carlson, who has over 25 years of experience in the biopharma industry.
Some of the team’s novel approaches include three computational pipelines that the Wyss recently developed to harness the power of data analytics, machine learning, and computer science address a number of diseases. These different approaches are now being deployed to rapidly evaluate existing drugs for potential activity against COVID-19.
The first uses a proprietary machine-learning algorithm called DRUID (DRUg Indication Discoverer) developed by senior staff scientist Diogo Camacho and his team. DRUID sifts through gene-expression data generated with tens of thousands of known drug compounds and identifies those that have potential to revert a disease-state expression pattern and phenotype back to a normal one. It was previously used to identify compounds with the potential to fight cancer and is now being used to analyze the gene-expression patterns of human lung cells infected with the CoV-2 virus, and how different drugs change those patterns.
The second approach, called molecular dynamics repurposing, was created by staff scientist Charles Reilly and his team. This uses multiscale computer-based molecular simulation techniques to create virtual versions of molecules whose properties can be modeled and analyzed. The team has used it to model the Spike protein found on the novel CoV-2 virus and develop small molecules that target a specific region of the protein. When tested in cultured cells infected with both pseudotyped and native CoV-2 viruses, some of these novel compounds inhibited infection in early studies. As part of the DARPA program, the team is now integrating this information with structural data from other drugs that inhibit CoV-2 infection to identify other FDA-approved drugs compounds that might have more potent effects.
The third, NemoCAD (network model for causality-aware discovery), created by senior engineer Richard Novak and his team, uses a network-analysis-based algorithm to compare genetic networks found in COVID-19 patients with those in healthy patients, and identifies drugs that could change their network state to that of a healthy patient. In past work, the team used this approach to identify existing approved drugs that could reversibly induce suspended animation in tadpoles or normalize behavioral abnormalities in a mouse model of Rett syndrome.
“There are so many great, creative scientific ideas happening all the time at the Wyss Institute, and bringing together these different projects into a unified process only makes them even stronger, because data about a given compound that is acquired using one method can then be shared with everyone else who is working on that compound from a different angle,” said Rani Powers, a senior staff scientist who is leading efforts to coordinate and integrate computational efforts and data relating to the therapeutics discovery program. “Each compound that we’re testing has a story that we’ll be able to follow, showing its journey from computational modeling to in vitro assays to animal models, and we’ll be able to see how the different assays performed on a compound provide a unique piece of the puzzle we’re trying to solve.”
From computers to organ chips to animal models
The Wyss cell biology team, including Ingber, senior staff scientist Rachelle Prantil-Baun, staff scientist Girija Goyal, and postdoctoral fellows Longlong Si and Haiqing Bai, recently uploaded a preprint to bioRxiv describing how they used cultured human lung cells in lung airway chips to identify the two approved compounds that inhibit infection with a CoV-2 pseudovirus at concentrations similar to those observed in human blood in clinical studies. But for any drug to be quickly approved for use in patients, it must be tested against the real CoV-2 virus. To make sure their candidates could continue their journey toward the clinic, Ingber teamed up with Frieman and tenOever, whose labs have dedicated spaces to safely conduct the tests, and together they created a full drug-testing pipeline that demonstrated the compounds’ safety and efficacy in animal models.
As part of the DARPA grant, Frieman and tenOever are also setting up organ chip testing programs in their own labs so they can infect human lung chips with the CoV-2 virus and study the inflammatory responses. The most active anti-CoV-2 compounds or drug combinations are being tested in tenOever’s CoV-2 animal models to validate efficacy, optimize dosing, and assess toxicity. Throughout the DARPA program, the team also will engage with other government partners and regulators to expedite the translation of drugs that prove effective inhibitors of CoV-2.
“Through our cell- and lung-on-a-chip-based antiviral testing system, we will be able to better predict candidate therapeutics for priority in animal models and eventually human trials,” said Frieman.
“The Wyss Institute always been a very collaborative institution, but addressing the COVID-19 crisis has required that we reach beyond our walls and beyond the immediate Boston community to identify partners who can build on our technological advances and add their own unique capabilities, much like handing off a baton to a teammate in a relay race, to achieve our shared goal of identifying existing drugs that can prevent this horrible disease,” said Ingber. “I’m confident that what we’re doing both internally and externally is going to help all of us cross the finish line together over the coming months.”





