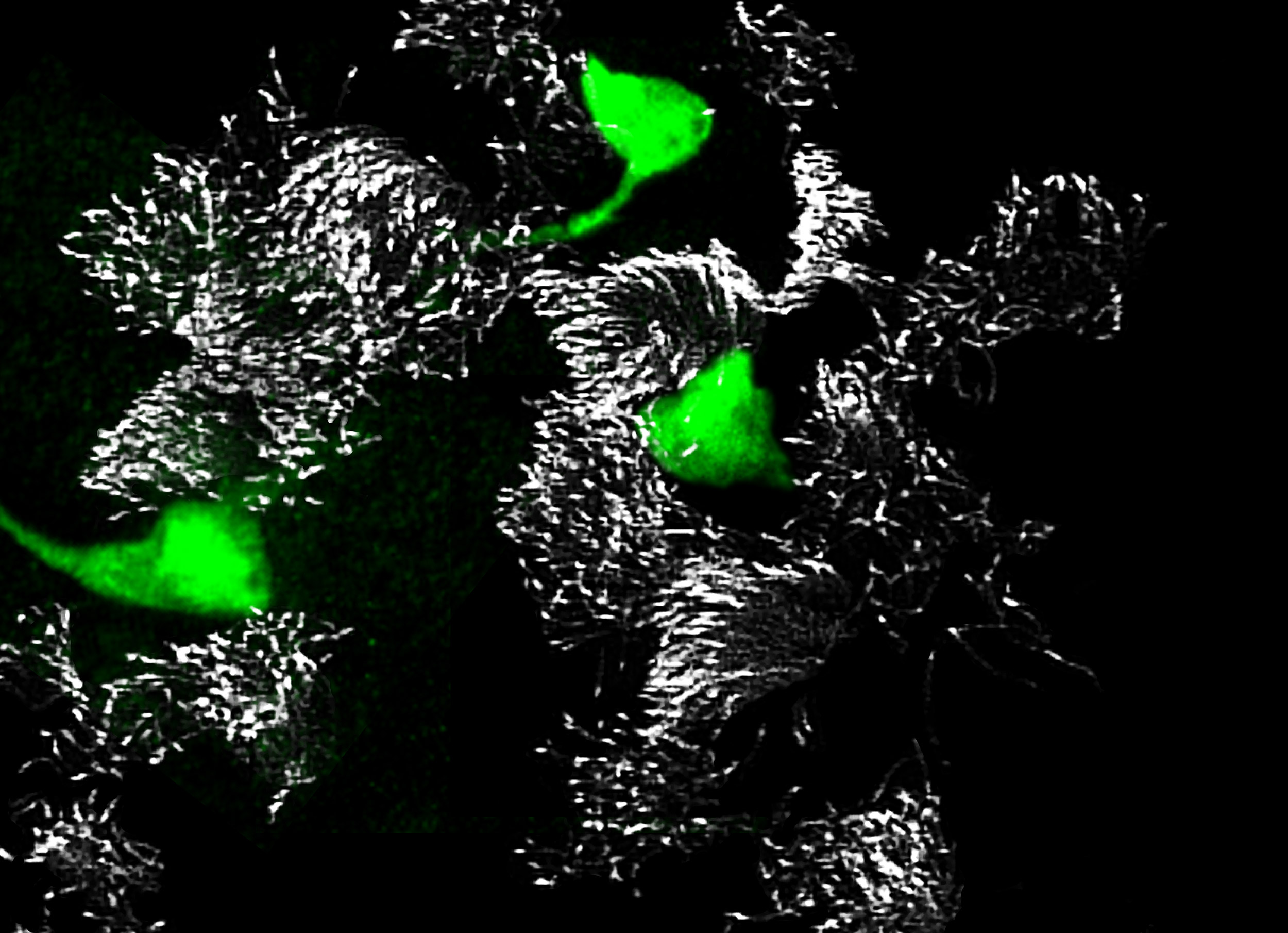
Newly identified, rare pulmonary ionocytes (green) dot the landscape of ciliated cells (white) of the mouse lung airway lining.
Credit: Montoro et al./Nature 2018
Cystic fibrosis clues found in newly identified cell type
These cells appear to be the primary source of activity of the gene responsible for multiorgan disease
In separate studies published online today in Nature, two independent research teams report the discovery of a new, rare type of cell in the human airway. These cells appear to be the primary source of activity of the CFTR gene, mutations which cause cystic fibrosis, a multiorgan disease that affects more than 70,000 people worldwide.
The CFTR gene provides instructions for making a protein called the cystic fibrosis transmembrane conductance regulator. This protein functions as a channel across the membrane of cells that produce mucus, sweat, saliva, tears, and digestive enzymes.
Despite decades of study on CFTR and progress in treating cystic fibrosis, there is still no cure. The new findings reveal that CFTR activity is concentrated in a small, previously unknown population of cells, which may serve as promising targets for future therapeutic strategies against the disease.
The researchers named the cells “pulmonary ionocytes” due to similarities with ionocytes, a type of cell that regulates salt balance and is found in the gills of freshwater fish and in frog skin.
One group was led by scientists from Harvard Medical School (HMS) and the Novartis Institutes for Biomedical Research (NIBR). The other group was led by HMS researchers based at Massachusetts General Hospital (MGH) and scientists at the Broad Institute of MIT and Harvard.
“As researchers work toward cures for cystic fibrosis, knowing you are looking at 1 percent of the cell population seems essential for any type of troubleshooting to improve a therapy or develop new therapies,” said Allon Klein, co-corresponding author of one of the Nature studies and assistant professor of systems biology at HMS.
The studies also revealed the characteristics of other new, rare, and poorly understood cell types, expanding the current understanding of lung biology and disease.
“Cystic fibrosis is an amazingly well-studied disease, and we’re still discovering completely new biology that may alter the way we approach it,” said Jayaraj Rajagopal, co-corresponding author of the second study and professor of medicine at MGH. “We have the framework now for a new cellular narrative of lung disease.”
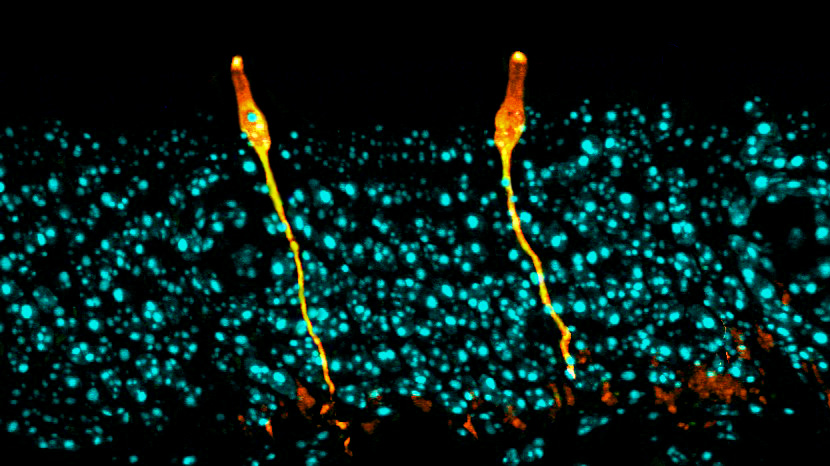
Single-cell surprise
Both teams set out to build an atlas of the cells that make up the airway. Using single-cell sequencing technology, they analyzed gene expression in tens of thousands of individual cells isolated from human and mouse airways — one cell at a time.
Comparing patterns of gene expression and using previously described cells as references, the researchers created comprehensive catalogs of different cell types and states, as well as their abundance and distribution.
The teams’ analyses mapped out the genetic identities of both known and previously undescribed cell types. The pulmonary ionocytes were particularly striking, as these cells expressed higher levels of CFTR than any others.
Identified in the late 1980s, CFTR (cystic fibrosis transmembrane conductance regulator) codes for a protein that transports chloride ions across cell membranes. Mutations to CFTR can lead to the buildup of thick mucus in the lungs, pancreas, and other organs, which in turn leads to frequent respiratory infections and other symptoms that characterize cystic fibrosis. Scientists have long assumed that CFTR is expressed at low levels in ciliated cells, a common airway cell type.
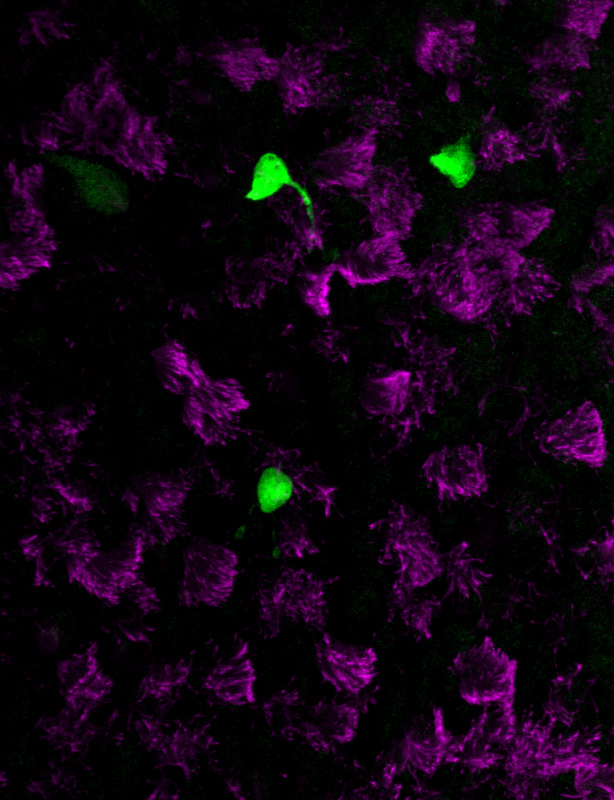
The new studies, however, suggest that the majority of CFTR expression occurs in pulmonary ionocytes, which make up only around 1 percent of airway cells. Klein, along with Aron Jaffe, a co-leader of respiratory disease research at NIBR, and colleagues additionally showed that the activity of CFTR, not just its expression, relates to the number of pulmonary ionocytes in the tissue.
“With single-cell sequencing technology, and dedicated efforts to map cell types in different tissues, we’re making new discoveries — new cells that we didn’t know existed, cell subtypes that are rare or haven’t been noticed before, even in systems that have been studied for decades,” said Broad core institute member and MIT professor of biology Aviv Regev, co-corresponding author of the study with Rajagopal.
When Regev, Rajagopal, and colleagues disrupted a critical molecular process in pulmonary ionocytes in mice, they observed the onset of key features associated with cystic fibrosis, notably the formation of dense mucus. This finding underscores how important these cells are to airway-surface regulation, the researchers said.
Together, the teams’ discoveries point to new strategies for treating cystic fibrosis, such as increasing the amount of pulmonary ionocytes to increase the amount of CFTR activity, said Jaffe, who is co-corresponding author of the study with Klein.
The identification of these cells can also help guide teams trying to use gene therapy to correct CFTR mutations, the authors suggest.
“We can use this information to be a bit more clever when we devise therapeutic approaches to cystic fibrosis,” Jaffe said.
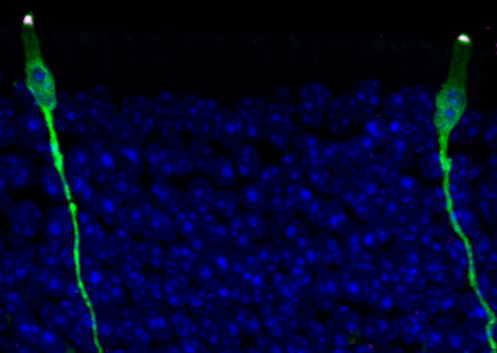
Here and rare
The teams’ analyses also shed light on new, rare, or poorly described cell states and subtypes, and characterized changes to certain cells after injury or during development.
Klein, Jaffe, and colleagues, for example, identified cell states that emerge or expand following damage or injury by studying regenerating tracheas. Using InDrops, a single-cell sequencing technology Klein and colleagues developed at HMS, at multiple points during regeneration, the team could track how cell states specific to the injury response transitioned over time.
Rajagopal, Regev, and colleagues developed a new method called Pulse-Seq to monitor development of cell types from their progenitors in the mouse airway, which were verified in human tissue. By adding a label to progenitor cells, they showed that mature cells in the airway arise from common progenitors known as basal cells.
A comprehensive atlas of airway cell types and their genetic fingerprints, both in normal conditions and in development and regeneration, now serves as potential baseline data for future studies of diseases and other health-related conditions, according to the research teams.
Rajagopal, Regev, and colleagues, for example, found that a gene linked to asthma development is specifically expressed by ciliated cells. Another gene linked with asthma is expressed in tuft cells, which separated into at least two groups — one that senses chemicals in the airway and another that produces inflammation. These findings may inform understanding and treatment of the disorder.
“We’ve uncovered a whole distribution of cell types that seem to be functionally relevant,” Rajagopal said. “What’s more, genes associated with complex lung diseases can now be linked to specific cells that we’ve characterized. The data are starting to change the way we think about lung diseases such as cystic fibrosis and asthma.”
Authors and funding
Lindsey Plasschaert, postdoctoral researcher at NIBR, and Rapolas Žilionis, graduate student at Vilnius University, Lithuania, and visiting scholar at HMS, are co-first authors, and Klein and Jaffe are co-corresponding authors of the study “A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte.”
Additional authors include Rayman Choo-Wing, Virginia Savova, Judith Knehr, and Guglielmo Roma. The study was supported by an award by the Burroughs Wellcome Fund, an Edward Mallinckrodt Jr. Foundation Grant, the Lithuanian Education Exchanges Support Foundation, and the National Cancer Institute (R33CA212697-01).
Daniel Montoro, graduate student at HMS, and Adam Haber and Moshe Biton, both postdoctoral fellows at the Broad, are co-first authors, and Rajagopal and Regev are co-corresponding authors of the study “A revised airway epithelial hierarchy includes CFTR-expressing ionocytes.”
Additional authors include Vladimir Vinarsky, Brian Lin, Susan Birket, Feng Yuan, Sijia Chen, Hui Min Leung, Jorge Villoria, Noga Rogel, Grace Burgin, Alexander Tsankov, Avinash Waghray, Michal Slyper, Julia Waldman, Lan Nguyen, Danielle Dionne, Orit Rozenblatt-Rosen, Purushothama Rao Tata, Hongmei Mou, Manjunatha Shivaraju, Hermann Bihler, Martin Mense, Guillermo Tearney, Steven Rowe, and John Engelhardt. The study was supported in part by the Klarman Cell Observatory at the Broad, the Manton Foundation, Howard Hughes Medical Institute, New York Stem Cell Foundation, the Harvard Stem Cell Institute, the Human Frontier Science Program, and the National Institutes of Health.
Author disclosures are listed in the respective studies.
Adapted from NIBR and Mass General/Broad Institute news releases





