
A team of researchers uses artificial intelligence technology to make it far easier than ever before to track animals’ movements in the lab.
Images courtesy of DeepLabCut
Movement monitor
An open-source AI tool for studying movement across behaviors and species
Understanding the brain, in part, means understanding how behavior is created.
To reverse-engineer how neural circuits drive behavior requires accurate and vigorous tracking of behavior, yet the increasingly complex tasks animals perform in the laboratory have made that challenging.
Now, a team of researchers from the Rowland Institute at Harvard, Harvard University, and the University of Tübingen is turning to artificial intelligence technology to solve the problem.
The software they developed, dubbed DeepLabCut, harnesses new learning techniques to track features from the digits of mice, to egg-laying behavior in Drosophila, and beyond. The work is described in an Aug. 20 paper published in Nature Neuroscience.
The software is the brainchild of Mackenzie Mathis, a Rowland Fellow at the Rowland Institute at Harvard; Alexander Mathis, a postdoctoral fellow working in the lab of Venkatesh N. Murthy, professor of molecular and cellular biology and chair of the Department of Molecular and Cellular Biology; and Matthias Bethge, a professor at the University of Tübingen and chair of the Bernstein Center for Computational Neuroscience Tübingen.
The notion of using software to track animal movements was born partly of necessity. Both Mackenzie and Alexander Mathis had tried using traditional techniques, which typically involve placing tracking markers on animals and using heuristics such as object segmentation, with mixed success.
Such techniques are often sensitive to the choice of analysis parameters, and markers or tattoos are invasive and can hinder natural behaviors, or may be impossible to place on very small or wild animals, they said.
Luckily, international competitions in recent years have driven advances in computer vision and the development of new algorithms capable of human pose-estimation (automatically tracking human body parts).
Such algorithms, however, are widely seen as data-hungry, requiring thousands of labeled examples for the artificial neural network to learn. This is prohibitively large for typical laboratory experiments, and would require days of manual labeling for each behavior.
The solution came in what is called “transfer learning,” or applying an already-trained network to a different problem, similar to the way scientists believe biological systems learn.
Using a state-of-the-art algorithm for tracking human movement called DeeperCut, the Mathises were able to show that deep learning could be highly data-efficient. The new software’s name is a nod to DeeperCut’s authors.
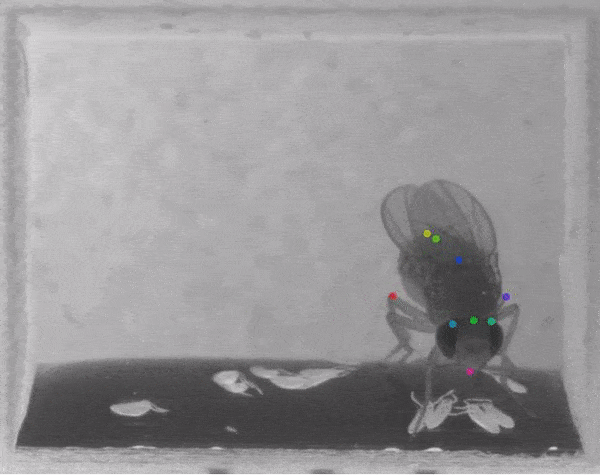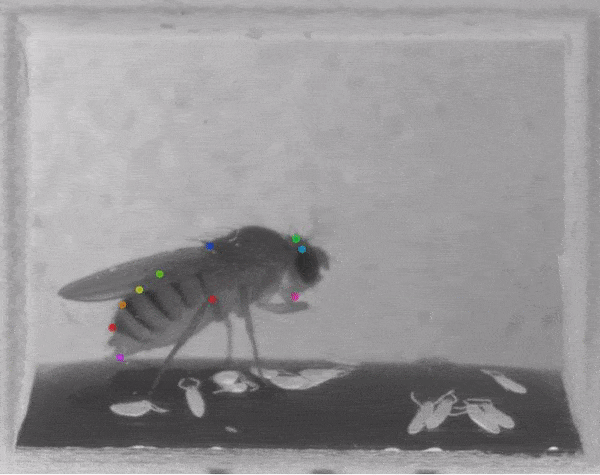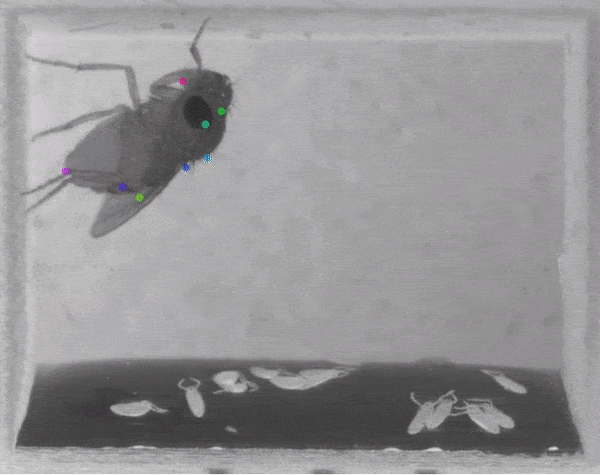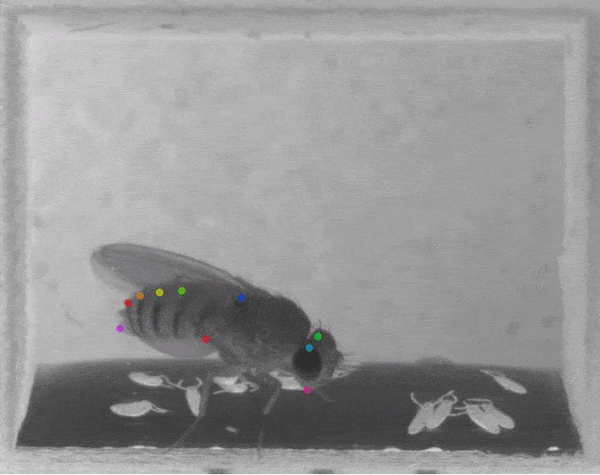
The software tracks the movements of a fly laying eggs and the digits of a mouse.
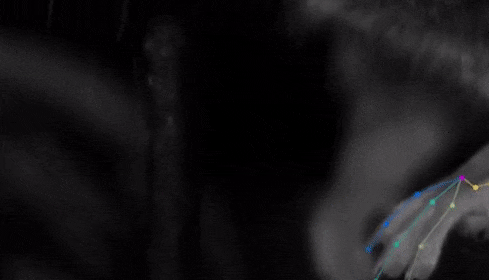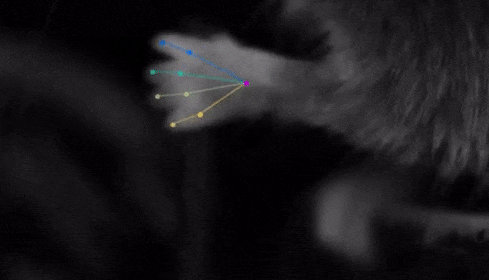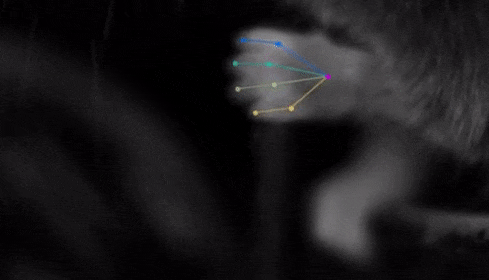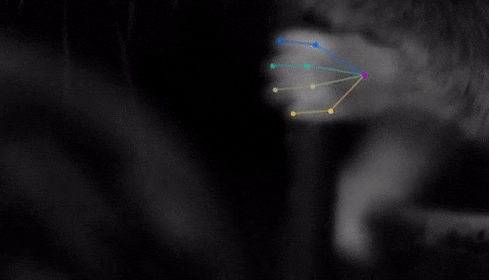
Just as a child does not need to develop another visual system from scratch in order to recognize a novel object, but relies on thousands of hours of experience and adapts them to recognize new objects, DeepLabCut is pretrained on thousands of images containing natural objects, images of hammers, cats, dogs, foods, and more.
With that pretraining in place, the software needed only 100 examples of mice performing an odor-guided navigation experiment to recognize specific mouse body parts as well as humans could.
The team was also able to apply the technology to mice making reaching movements, and, in collaboration with Kevin Cury, a neuroscientist from Columbia University, to flies laying eggs in a 3-D chamber.
“We were very impressed by the success of the transfer-learning approach and the versatility of DeepLabCut,” Mackenzie Mathis said. “With only a few hundred frames of training data, we were able to get accurate and robust tracking across a myriad of experimental conditions, animals, and behaviors.”
“Experimentalists have very good intuitions about what body parts should be analyzed to study a particular behavior, but traditionally extracting limb coordinates from videos has been very challenging — DeepLabCut does just that based on a few examples,” Alexander Mathis said. “Since the program is designed as a user-friendly, ‘plug-and-play’ solution, and does not require any coding skills, it can be widely used.”
“We want as many researchers as possible to benefit from our work,” said Bethge. “DeepLabCut was created as an open software, as sharing results, data, and also algorithms is essential for scientific progress.”
Even as the paper describing the software was published, the technology had been used by more than 50 labs to study everything from the gait of horses to bacteria dynamics to the movement of surgery robots.
The software toolbox can be used with minimal to no coding experience and is freely available at mousemotorlab.org/deeplabcut.
This study was supported with funding from the Marie Sklodowska-Curie International Fellowship, the Rowland Institute at Harvard, Project ALS Women & the Brain Neuroscience Fellowship, the German Science foundation (DFG) CRC 1233 on Robust Vision, and IARPA through the MICrONS program.





