Single-stranded DNA and RNA origami go live
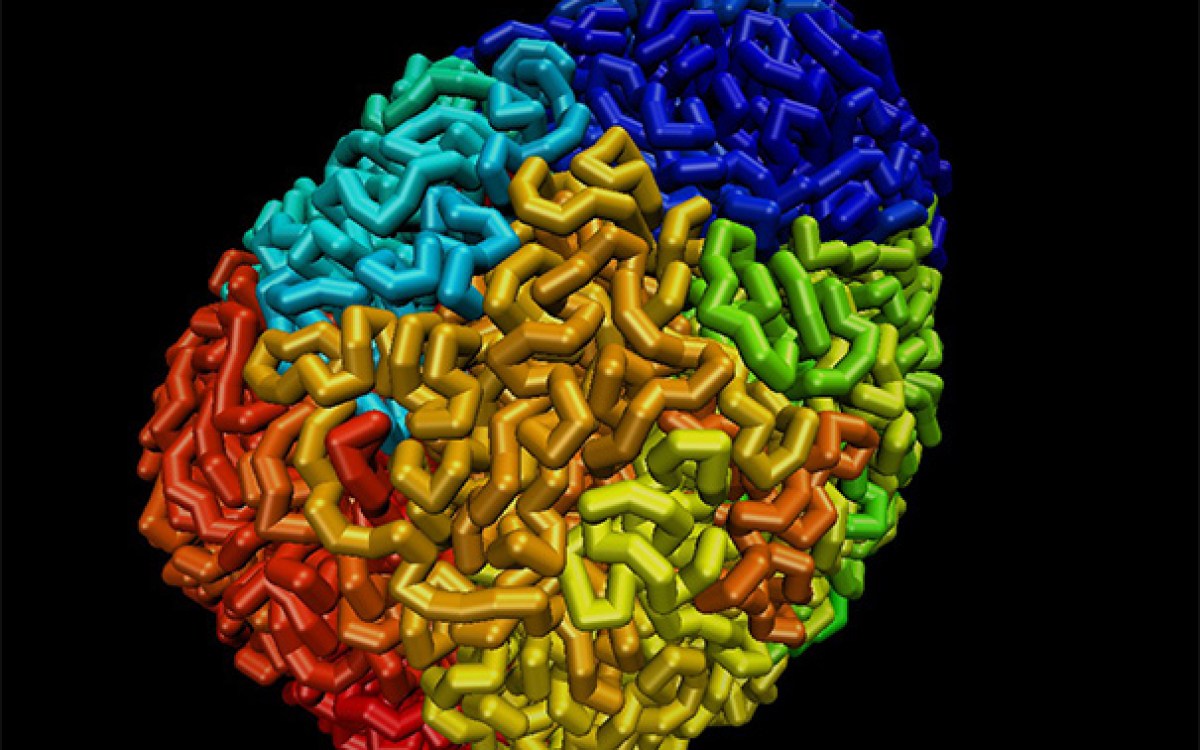
Credit: Molgraphics
New approach has potential to create tiny nanofactories for production of pharmaceuticals
Nanotechnologists are using DNA, the genetic material present in living organisms, as well as its multifunctional cousin RNA as the raw material in efforts to build minuscule devices that could potentially function as drug-delivery vehicles, tiny nanofactories that produce pharmaceuticals and chemicals, or highly sensitive elements of electric and optical technologies.
Like genetic DNA (and RNA) in nature, these engineered nanotechnological devices are also made up of strands comprised of the four bases known in shorthand as A, C, T, and G. Regions within those strands can spontaneously fold and bind to each other via short, complementary base sequences in which As from one sequence bind to Ts from another, and Cs to Gs.
Researchers at the Wyss Institute of Biologically Inspired Engineering at Harvard University and elsewhere have used these features to design self-assembling nanostructures such as scaffolded DNA origami and DNA bricks of ever-growing sizes and complexities that are becoming useful for diverse applications. However, the translation of these structures into medical and industrial applications is still challenging, partially because these multistranded systems are prone to local defects due to missing strands. In addition, they self-assemble from hundreds to thousands of individual DNA sequences that each must be verified and tested for high-precision applications, and whose expensive synthesis often produces undesired side products.
Now, a novel approach published in Science by a collaborative team of researchers from the Wyss Institute, Arizona State University, and Autodesk for the first time enables the design of complex, single-stranded DNA and RNA origami that can autonomously fold into diverse, stable, user-defined structures. In contrast to the synthesis of multistranded nanostructures, these entirely new types of origami are folded from a single strand, which can be replicated in living cells. This could allow for low-cost production at large scales and with high purities, opening entirely new opportunities for diverse applications such as drug delivery and nanofabrication.
Earlier generations of larger-sized origami were composed of a central scaffold strand whose folding and stability required more than 200 short staple strands that bridged distant parts of the scaffold and fixed them in space. “In contrast to traditional scaffolded origamis, which are assembled from hundreds of components, our new approach allows us to reliably design and synthesize stable single-stranded and self-folding origami,” said Wyss Institute core faculty member and corresponding author Peng Yin. “Our fundamentally new approach relies on single-strand folding, rather than multicomponent assembly, to produce large nanostructures. This, together with the ability to basically clone and multiply the single component strand in bacteria, presents a game-changing advance in DNA nanotechnology that greatly enhances single-stranded origami’s potential for real-world applications.” Yin is also co-lead of the Wyss Institute’s Molecular Robotics Initiative and professor of systems biology at Harvard Medical School (HMS).
To enable the production of single-stranded and stable DNA-based origami with distinct folding patterns, the team first had to overcome several challenges. In a large DNA strand that goes through a complex folding process, many sequences need to accurately pair up with far-away sequences. If this process does not happen in an orderly and precise fashion, the strand gets tangled and forms unspecific knots along the way, rendering it useless.
“To avoid this problem, we identified new design rules that we can use to cross DNA strands between different double-stranded regions, and developed a web-based automated design tool that allows researchers to integrate many of these events into a folding path leading up to a large, knot-free nanocomplex,” said Dongran Han, the study’s first author and a postdoctoral fellow on Yin’s team.
The largest DNA origami structures created previously were assembled by synthesizing all their constituent sequences individually in vitro and mixing them together. As a key feature of the new design process, the single-strandedness of the DNA origami allowed the researchers to introduce DNA sequences stably into E. coli bacteria to inexpensively and accurately replicate them with every cell division. “This could greatly facilitate the development of single-stranded origami for high-precision nanotech like drug delivery vehicles, for example, as only a single, easy-to-produce molecule needs to be validated and approved,” said Han.
Finally, the team also adapted single-stranded origami technology to RNA, which, as a different nucleic acid material, offers certain advantages, such as even higher production levels in bacteria, and usefulness for potential intracellular and therapeutic RNA applications. Translating the approach to RNA scales up the size and complexity of synthetic RNA structures 10-fold compared to previous structures made from RNA.
The proof-of-concept analysis also proved that protruding DNA loops can be precisely positioned and be used as handles to attach functional proteins. In future developments, single-stranded origami thus could be functionalized by attaching enzymes, fluorescent probes, metal particles, or drugs either to their surfaces or within cavities inside. This could effectively convert single-stranded origami into nanofactories, light-sensing and -emitting optical devices, or drug-delivery vehicles.
Read the full press release.
The study was funded by the Office of Naval Research, the Army Research Office, the National Science Foundation’s Expeditions in Computing program, and the Wyss Institute for Biologically Inspired Engineering.