“Turing was brilliant,” said Alexander Schier, professor of molecular and cellular biology. “There wasn’t a single molecule known that would regulate development or pattern formation when he proposed this model. For him, it was a pure mathematical model.
File photo by Stephanie Mitchell/Harvard Staff Photographer
Turing was right
Two proteins fit decades-old prediction
Today, Alan Turing is best known as the father of modern computer science, but in 1952 he sketched out a biological model in which two chemicals — an activator and an inhibitor — could interact to form the basis for everything from the color patterns of a butterfly’s wings to the black and white stripes of a zebra.
It was an innovative hypothesis, made all the more impressive by the fact that it was postulated without the benefit of modern molecular biology — the double-helix structure of DNA wouldn’t be discovered for another year.
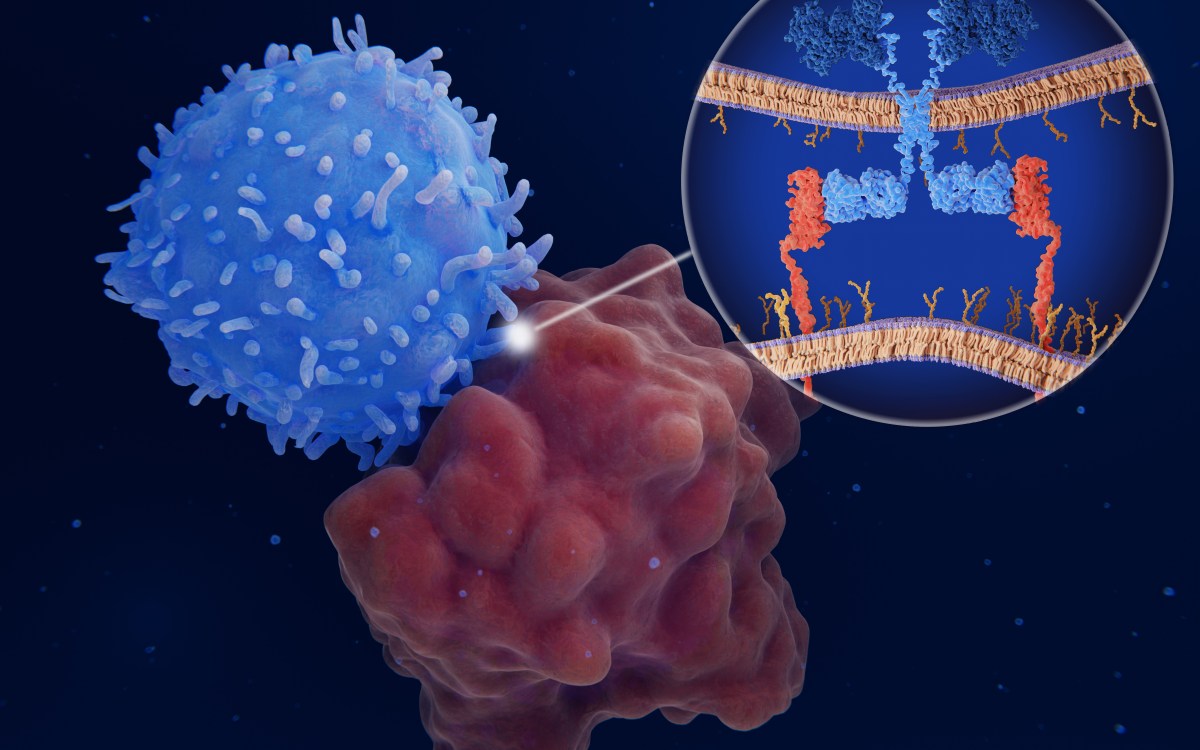
Harvard research now shows that Nodal and Lefty — two proteins linked to the regulation of asymmetry in vertebrates and the development of precursor cells for internal organs — fit the model described by Turing six decades ago. In a paper published online in Science April 12, Alexander Schier, professor of molecular and cellular biology, and his collaborators Patrick Müller, Katherine Rogers, Ben Jordan, Joon Lee, Drew Robson, and Sharad Ramanathan demonstrate a key aspect of Turing’s model: that the activator protein Nodal moves through tissue far more slowly than its inhibitor Lefty.
“That’s one of the central predictions of the Turing model,” Schier said. “So I think we can now say that Nodal and Lefty are a clear example of this model in vivo.”
Schier’s latest finding is the result of more than a decade of research into the Nodal/Lefty pairing. In a 2002 paper, his group described results that suggested the two proteins act as an activator/inhibitor pair, one of the key tenets of the model outlined by Turing. But it was the recent experiments on how the proteins move through tissue that clinched it.
To test the biophysical properties of Nodal and Lefty, Schier’s team began by generating modified versions of Nodal or Lefty that would fluoresce under laser light. They observed that Lefty moved farther through zebrafish embryos than Nodal.
To measure the diffusion rates of these proteins, they used a process called photobleaching to “erase” an area of either Nodal or Lefty. They then measured the time needed for Lefty and Nodal to diffuse into the bleached space. The results matched the prediction of the Turing model.
In a separate test, the researchers explored whether the two proteins might have different stabilities, which could also explain why Lefty moves farther through the embryo than Nodal. To test this possibility, researchers irradiated the modified versions of either Nodal or Lefty with lasers, causing the fluorescent proteins to change their color from green to red. By measuring how long it took for the red color to disappear, they were able to determine that Nodal and Lefty are similarly stable.
“That tells us that it’s the mobility not the stability that is different between these two molecules,” Schier said. “That’s important, because it is the diffusion that’s different in the models proposed by Turing.
“Turing was brilliant,” Schier continued. “There wasn’t a single molecule known that would regulate development or pattern formation when he proposed this model. For him, it was a pure mathematical model. The Turing equation is simple but there’s a certain beauty to it. It can be applied to many different biological systems and what you get are amazing and beautiful patterns. Our paper shows that aspects of the Turing model actually do work in vivo. We still don’t know how a zebra’s stripes or a leopard’s spots are formed, but the Turing model shows one way it could work.”
Going forward, Schier said, he hopes to understand the mechanism behind the different mobility of Nodal and Lefty. “We know these proteins are different, but why are they different?” Schier asked. “They are similar in size, they have similar structures – but somehow they must interact differently with other molecules, affecting how they move. That’s a question for the future.”