Photo by Eliza Grinnell/SEAS
Brain navigation
Research team turns terabytes of image data into model of neural circuits
The brain of a mouse measures only 1 cubic centimeter in volume. But when neuroscientists at Harvard’s Center for Brain Science slice it thinly and take high-resolution micrographs of each slice, that tiny brain turns into an exabyte of image data. That’s 1018 bytes, equivalent to more than a billion CDs.
What can you do with such a gigantic, unwieldy data set? That’s the latest challenge for Hanspeter Pfister, the Gordon McKay Professor of the Practice of Computer Science at Harvard’s School of Engineering and Applied Sciences (SEAS).
Pfister, an expert in high-performance computing and visualization, is part of an interdisciplinary team collaborating on the Connectome Project at the Center for Brain Science. The project aims to create a wiring diagram of all the neurons in the brain. Neuroscientists have developed innovative techniques for automatically imaging slices of mouse brain, yielding terabytes of data so far.
Pfister’s system for displaying and processing these images would be familiar to anyone who has used Google Maps. Because only a subsection of a very large image can be displayed on a screen, only that viewable subsection is loaded. Drag the image around, zoom in or out, and more of the image is displayed on the fly.
This “demand-driven distributed computation” is the central idea behind Pfister’s work, for which he recently won a Google Faculty Research Award.
With new techniques in science producing ever-larger data sets, the problem is hardly unique to the Connectome Project. Pfister is certain that the tools his research group develops will be useful to scientists working with large image data sets in any field — for example, astronomers processing radio telescope images or Earth scientists analyzing atmospheric data.
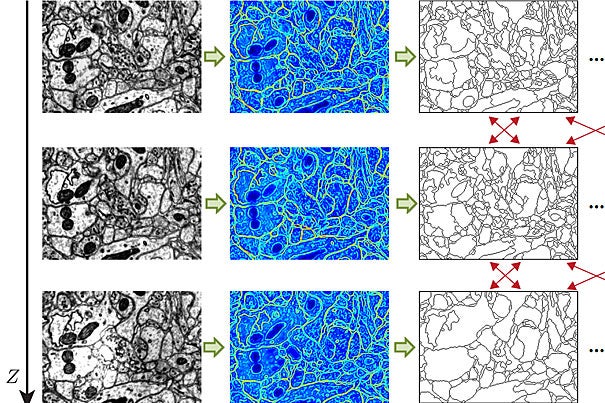
For the Connectome, the ultimate tool will be much more than “Google Maps for the brain.” The project has two crucial new features, says Pfister. First, the mouse brain must be reconstructed in three dimensions, so one can quickly flip through the stack of images in addition to zooming in on and panning across it. The images, each representing a 30-nanometer-thick brain slice, can also be stacked together and viewed from the side, along the same axis they were sliced.
Additionally, the system developed by Pfister’s research team extends the demand-driven principle to image analysis and processing. If a neuroscientist wants to automatically align images, adjust the brightness of an image, or run an automatic cell boundary detector over a portion of the images, for example, these operations are also computed on the fly, so there is no need to wait for the entire stack of images to be processed before the relevant subsection is viewable. Pfister compares it with “a very specialized version of Adobe Photoshop software.”
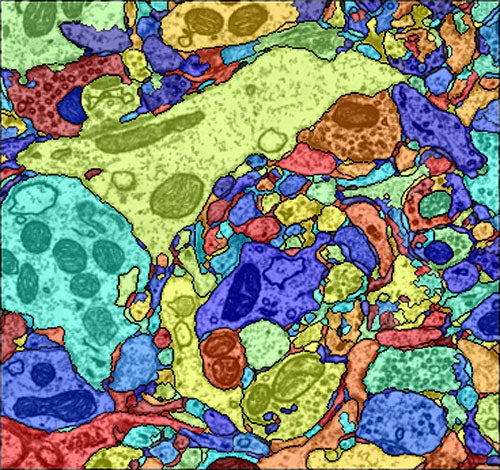
Indeed, the existing tools are capable of far more advanced image processing. Since the collaboration began in 2007, computer scientists in Pfister’s group have created algorithms to automatically detect cell boundaries, a process known as segmentation, and to connect related cells in three dimensions, a process known as reconstruction. Previously, neuroscientists did this by hand.
With thousands of neurons in even a cubic millimeter of brain tissue, it would be impossible to scale the project up without automated segmentation, says Jeff Lichtman, the Jeremy R. Knowles Professor of Molecular and Cellular Biology at Harvard and leader of the Connectome Project.
Automated segmentation is done by a machine-learning algorithm, which a neurobiologist trains by marking the cell boundaries in as few as five images. The algorithm then learns the features — edges, corners, textures, etc. — associated with cell boundaries. Occasional errors can be corrected on the go, and the algorithm learns from these errors. With all the cell boundaries in a stack of images identified, a second machine-learning method combines information from the images to reconstruct the shape of a neuron in three-dimensional space.
Researchers in Pfister’s group are working on ways to automatically detect synapses, the sites where two neurons are connected.
“We spent four years developing algorithms to find cell boundaries and synapses as well as building up the infrastructure for viewing and displaying large image data sets,” says Pfister. The individual tools are in place, but “now we’re putting the two pieces together,” he adds, looking ahead to the next few years of research.
The Connectome Project is certainly an ambitious one. The sheer quantity of electron microscopy slices to be acquired and analyzed is a daunting but necessary challenge, says Lichtman. “We have made no progress on simple questions like how many neurons converge on a single neuron. A lot of brain functions are at that level.”
Lichtman believes that the Connectome will be able to answer those simple questions and also complex ones. The patterns that emerge about how different types of neurons connect to one another can give physical clues to how memories and personality traits are encoded. That is all far off, but it would be impossible without the analysis tools developed by computer scientists.
The Connectome Project has drawn together collaborators in neurobiology, molecular and cellular biology, medicine, and chemistry and chemical biology.
“This is a great example of a cross-disciplinary project,” said Pfister. “At SEAS we are a bridge between different Schools, which makes it easy for me to work with my collaborators. It’s very gratifying to see our tools used in real applications.”